-Search query
-Search result
Showing 1 - 50 of 211 items for (author: rosenthal & p)

EMDB-17001: 
In-situ structure of the hexameric HEF trimers from influenza C viral particles
Method: subtomogram averaging / : Liu ZB, Rosenthal PB
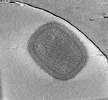
EMDB-18916: 
Cryotomogram of mature Vaccinia virus (WR) virion
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18917: 
Subtomogram average of the Vaccinia virus (WR) portal complex in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18918: 
Subtomogram average of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

PDB-8r5i: 
In situ structure of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions by flexible fitting into a cryoET map
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-16258: 
Negative stain EM structure of Toxoplasma gondii glideosome-associated connector (subdomain coil 1-3)
Method: single particle / : Chen Q, Rosenthal P

EMDB-16259: 
Negative stain EM structure of Toxoplasma gondii glideosome-associated connector subdomain coil 3
Method: single particle / : Chen Q, Rosenthal P

EMDB-16260: 
Negative stain EM structure of Toxoplasma gondii glideosome-associated connector (subdomain coil 1-2)
Method: single particle / : Chen Q, Rosenthal P

EMDB-16257: 
Cryo-EM structure of Toxoplasma gondii glideosome-associated connector
Method: single particle / : Chen Q, Rosenthal P

EMDB-16670: 
Structure of the SNV L protein bound to 5' RNA
Method: single particle / : Meier K, Thorkelsson SR, Durieux Trouilleton Q, Vogel D, Yu D, Kosinski J, Cusack S, Malet H, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8ci5: 
Structure of the SNV L protein bound to 5' RNA
Method: single particle / : Meier K, Thorkelsson SR, Durieux Trouilleton Q, Vogel D, Yu D, Kosinski J, Cusack S, Malet H, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-16150: 
8:1 binding of FcMR on IgM pentameric core
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-16151: 
FcMR binding at subunit Fcu1 of IgM pentamer
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-16152: 
FcMR binding at subunit Fcu3 of IgM pentamer
Method: single particle / : Chen Q, Rosenthal P, Tolar P

PDB-8bpe: 
8:1 binding of FcMR on IgM pentameric core
Method: single particle / : Chen Q, Rosenthal P, Tolar P

PDB-8bpf: 
FcMR binding at subunit Fcu1 of IgM pentamer
Method: single particle / : Chen Q, Rosenthal P, Tolar P

PDB-8bpg: 
FcMR binding at subunit Fcu3 of IgM pentamer
Method: single particle / : Chen Q, Rosenthal P, Tolar P

PDB-8cco: 
JAS-stabilized F-ActinII from Plasmodium falciparum
Method: helical / : Kursula I, Lopez AJ

EMDB-15596: 
In situ subtomogram average of Vaccinia virus (WR) palisade, all virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15597: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from CEVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15598: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from IMVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15599: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from IEVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15600: 
Cryotomogram of Vaccinia virus (WR) infected HeLa cell (immature virions)
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15601: 
Cryotomogram of Vaccinia virus (WR) infected HeLa cell (mature virions)
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15602: 
In situ subtomogram average of Vaccinia virus (WR) D13 lattice, on immature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

PDB-8arh: 
In situ subtomogram average of Vaccinia virus (WR) D13 lattice, on immature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15607: 
Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15608: 
Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15610: 
Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15614: 
Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15615: 
Structure of the SFTSV L protein bound in a resting state [RESTING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8as6: 
Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8as7: 
Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8asb: 
Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8asd: 
Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8asg: 
Structure of the SFTSV L protein bound in a resting state [RESTING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15181: 
Prefusion spike of SARS-CoV-2 (Wuhan), closed conformation
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB
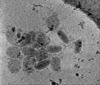
EMDB-15182: 
Electron cryotomography of SARS-CoV-2 virions
Method: electron tomography / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-15183: 
Whole SARS-CoV-2 (Wuhan) virion
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-15185: 
Prefusion spike of SARS-CoV-2 (Wuhan), 1-RBD-up conformation
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-13921: 
Cryo-EM structure of full-length human immunoglobulin M
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-13922: 
Cryo-EM structure of human monomeric IgM-Fc
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15375: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 1
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15376: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 2
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15377: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 3
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15379: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 5
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15380: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 4
Method: single particle / : Chen Q, Rosenthal P, Tolar P

PDB-7qdo: 
Cryo-EM structure of human monomeric IgM-Fc
Method: single particle / : Chen Q, Rosenthal P, Tolar P

PDB-8ady: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 1
Method: single particle / : Chen Q, Rosenthal P, Tolar P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
